Data Viewer#
Important
This section assumes that the reader is already familiar with Application Settings.
Introduction#
By default, the data viewer in specvizitor is configured to display the Grizli data products. Specifically, it is loading data from the following files:
File |
Description |
|---|---|
|
1D spectrum |
|
2D spectrum stack of all exposures |
|
Additional products including emission line maps and image cutouts |
However, data products handled by specvizitor are not limited to this list. By modifying the data_widgets.yml file, you can create a custom widget configuration tailored to practically any spectroscopic dataset. In this section, you will learn how to configure data widgets in specvizitor, starting with making small tweaks to the default configuration and finishing with writing a configuration “from scratch”.
Configuring the default widgets#
Let us start with some examples of how you can configure the default data widgets.
Changing the redshift range#
In data_widgets.yml, navigate to plots ‣ Spectrum 1D ‣ redshift_slider ‣ max_value:
...
plots:
Spectrum 1D:
...
redshift_slider:
...
max_value: 10
Set max_value to any redshift you think would suffice to classify even the most distant objects included in your sample. Next, make the same changes to the redshift slider under images ‣ Spectrum 2D:
images:
...
Spectrum 2D:
...
redshift_slider:
...
max_value: 10
This is required because Spectrum 1D and Spectrum 2D share the same redshift. Once you have made the changes, save data_widgets.yml and launch specvizitor. The maximum value of the redshift slider should be updated accordingly.
Changing the color bar range#
Note
This example demonstrates how to change the color bar range of Spectrum 2D, however the same applies to any widget in the images category.
In data_widgets.yml, navigate to images ‣ Spectrum 2D ‣ color_bar ‣ limits:
images:
...
Spectrum 2D:
...
color_bar:
...
limits:
min: -0.015
max: 0.015
type: user
Here, you can set the min and max parameters of the color bar. Once you have made the changes, save data_widgets.yml and launch specvizitor. The color bar range in Spectrum 2D should be updated accordingly.
Linking widget elements#
Note
This example demonstrates how to link plot axes, however the same applies to sliders and color bars.
In data_widgets.yml, navigate to images ‣ Line Map 1 ‣ x_axis ‣ link_to:
images:
...
Line Map 1:
...
x_axis:
link_to: null
Set link_to to Image Cutout. Once you have made the changes, save data_widgets.yml and launch specvizitor. The y-axes of Image Cutout and Line Map 1 should be linked together.
Hiding widget elements#
Note
This example demonstrates how to change the visibility of plot axes, however the same applies to color bars, sliders, spectral lines, and widgets themselves.
Tip
Most of the widget elements can be hidden from the UI by pressing H.
In data_widgets.yml, navigate to images ‣ Spectrum 2D ‣ x_axis ‣ visible:
images:
...
Spectrum 2D:
...
x_axis:
visible: true
Set visible to false, save data_widgets.yml and launch specvizitor. The Spectrum 2D’s x-axis will disappear from the view.
Adding new widgets#
Let us continue with some examples of how you can add new widgets to the data viewer.
Widget types#
There are two types of widgets that can be added to the data viewer — images and plots:
images:
...
plots:
...
Both images and plots can include multiple items:
images:
Image:
...
Spectrum 2D:
...
plots:
Plot:
...
Plot 2:
...
Another Plot:
...
Important
All widget names must be unique.
The difference between images and plots lies in the parameters that widgets of these types can have. For example, the color_bar parameter is unique to widgets listed in images, and the plots parameter is unique to widgets listed in plots.
Adding an image#
This is a minimal example of data_widgets.yml with a single image configuration:
images:
Image Cutout:
data:
filename: '{root}_{id:05d}.stack.fits'
Sometimes a FITS file contains multiple images (tables). By default, specvizitor loads the first image (table) from such files. To load a different image (table), we can specify the extname and extver parameters in loader_params:
images:
Image Cutout:
data:
filename: '{root}_{id:05d}.stack.fits'
loader_params:
extname: SCI
extver: F356W
In addition, it is often useful to configure the default range of the color bar displayed next to the image:
images:
Image Cutout:
data:
filename: '{root}_{id:05d}.stack.fits'
loader_params:
extname: SCI
extver: F356W
color_bar:
limits:
type: user
min: -0.015
max: 0.015
Here, type mimics the types of images limits used in SAOImage DS9 and can be either user (in which case it is necessary to specify min and max), minmax (the default value), or zscale.
Adding a plot#
Similarly to images, this is a minimal example of data_widgets.yml with a single plot configuration:
plots:
Spectrum 1D:
data:
filename: '{root}_{id:05d}.1D.fits'
This is how we can specify which plot(s) will be shown in this widget:
plots:
Spectrum 1D:
data:
filename: '{root}_{id:05d}.1D.fits'
plots:
flux:
x: wave
y: flux
Here, wave and flux refer to the columns of the table loaded from the FITS file.
In addition, we might want to configure the plot limits:
plots:
Spectrum 1D:
data:
filename: '{root}_{id:05d}.1D.fits'
plots:
flux:
x: wave
y: flux
y_axis:
limits:
min: -0.5
max: 1.5
Finally, let us add a redshift slider to the widget and make spectral lines visible in the plot:
plots:
Spectrum 1D:
data:
filename: '{root}_{id:05d}.1D.fits'
plots:
flux:
x: wave
y: flux
y_axis:
limits:
min: -0.5
max: 1.5
redshift_slider:
visible: true
max_value: 10
step: 1.0e-6
catalog_name: redshift
show_text_editor: true
show_save_button: true
spectral_lines:
visible: true
A full example#
This is a full example of data_widgets.yml which combines the two previous examples:
images:
Image Cutout:
data:
filename: '{root}_{id:05d}.stack.fits'
loader_params:
extname: SCI
extver: F356W
color_bar:
limits:
type: user
min: -0.015
max: 0.015
plots:
Spectrum 1D:
data:
filename: '{root}_{id:05d}.1D.fits'
plots:
flux:
x: wave
y: flux
y_axis:
limits:
min: -0.5
max: 1.5
redshift_slider:
visible: true
max_value: 10
step: 1.0e-6
catalog_name: redshift
show_text_editor: true
show_save_button: true
spectral_lines:
visible: true
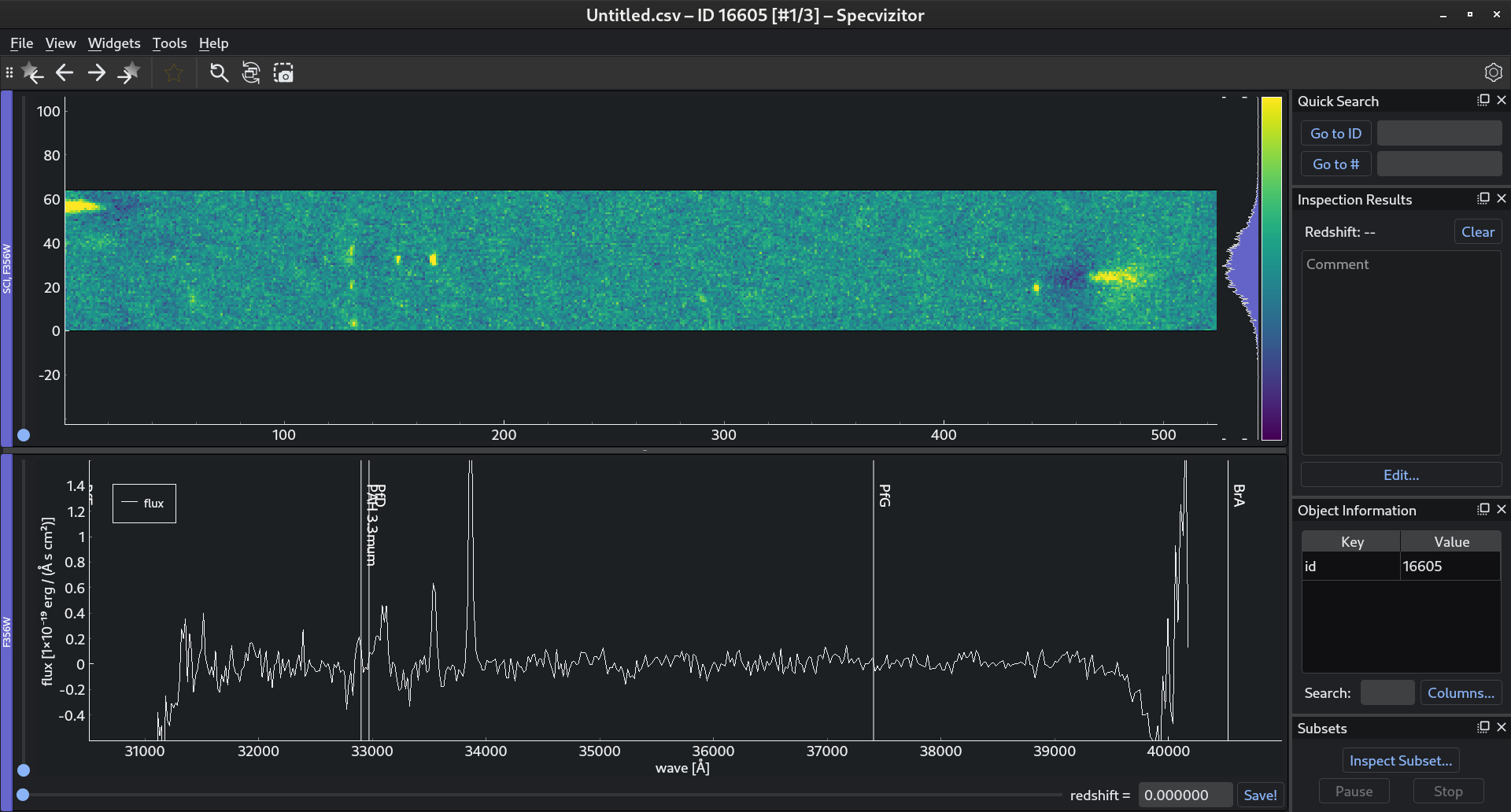
If you load this file in specvizitor (), you should see the following:
Tip
The default widget configuration is available here.